Population variation in gut microbiota
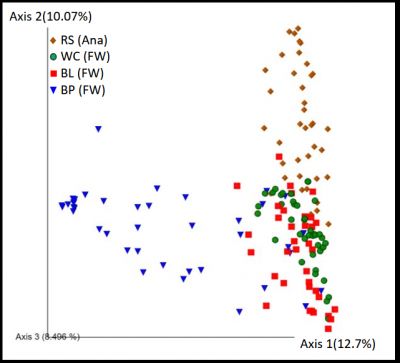
Individuals within the same species can have different gut microbiota, but little is known about the variation in gut microbiota between populations. We have snapshots of gut microbiota from 4 different Alaskan populations that shows that there is greater variation between the populations of stickleback compared to within a population, and that some populations have similar microbiota (figure by Ryan Lucas, 2020). To determine whether these differences change over time and what factors may drive those changes, we are currently analyzing the gut microbiota populations that were sampled annually for 8 years (4 populations) or monthly for 3 years (1 population). We are also part of a huge ecology-evolution experiment led by Andrew Hendry of McGill University in which stickleback fish were transplanted from various lakes in Alaska and put into new lakes in 2019, and then sampled annually to follow the evolution of those fish. While our lab is characterizing the gut microbiota, other labs are characterizing other aspects including diet, parasite infection rates, metabolism, immune system development, genetics, and environmental factors like prey, water chemistry, and more. All these experiments will help us determine what factors drive selection for different microbiota communities and how these relationships change over time.
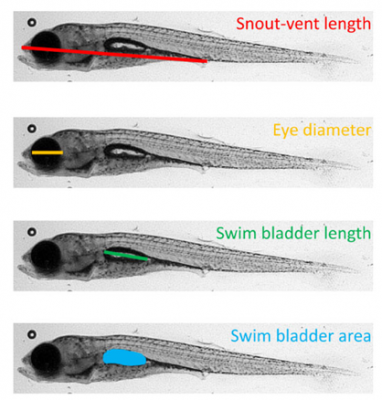
Effect of the microbiiota on stickleback development
We have a library of over 400 microbes isolated from stickleback guts from various populations and life stages. We also have a protocol to generate fish germ free, meaning they have no bacteria associated with them, and to maintain them germ free for the first couple of weeks. Since stickleback fish are transparent their first few weeks of life, we can use the germ free protocol and our library of microbes to determine how individual microbes or communities of microbes affect the internal and external development of the fish in real time. We have determined that the gut microbiota of stickleback affect the development of the host different depending on the population of fish, and that different pathways are affected different depending on both the population and the microbes examined. We are now in the process of using our sequencing results to identify specific microbes to characterize further, and comparing in vitro interactions to in vivo interactions to determine how the host affects microbe-microbe interactions, and whether that varies between populations. Image from Kirshman 2020.
Effects of ecotoxicants on host development
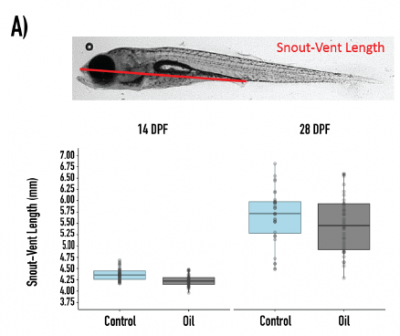
Dr. Kat is from Alaska. While most people think of Alaska waters as being pristine, we have ample evidence that human-made contaminants have affected the environment in negative ways. We are using our library of stickleback microbes to determine whether any of our microbes can breakdown common Alaska contaminants, and whether we can use these to protect fish against these contaminants. Thus far, we have examined how perchlorate (with Frank von Hipple of the University of Arizona), crude oil (with Mary Beth Leigh at the University of Alaska Fairbanks and Brandon Briggs at the University of Alaska Anchorage), fluridone (with Pat Tomco at the University of Alaska Anchorage), antibiotics, and microplastics are affected by stickleback microbes and how these affect the microbiota of stickleback fish. We are now interested in how in vitro microbe-microbe interactions affect ecotoxicant breakdown, and whether these microbes can protect the host against the ecotoxicants, and whether these interactions vary between populations of stickleback fish. Image from Ireland BioRxiv 2021.